Load model and data files
Load the tree inference model and example data available within
treepplr.
The data in this example is a toy dataset …
str(data)
#> List of 1
#> $ data: int [1:4, 1:15] 1 1 0 0 1 1 0 0 0 0 ...Run TreePPL
Now we can compile and run the TreePPL program. The function
tp_treeppl() has many optional arguments to change the
inference method used. Here, we will use
output_list <- tp_treeppl(model = model, data = data, method = "smc-apf",
samples = 1000, subsample = 5, resample = "manual",
n_runs = 10)The result is a list of sweeps, each one containing 3 elements: the sampled parameter values and the weights (in log scale) for each of the 5 particles in each sweep, and the normalizing constant for the whole sweep.
str(output_list,max.level = 2)
#> List of 10
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.1
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.3
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.4
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.3
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.5
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.4
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.4
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.4
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.5
#> $ :List of 3
#> ..$ samples :List of 10
#> ..$ weights :List of 10
#> ..$ normConst: num -79.4Plot the posterior distribution
In this example we will check that the different runs converged to the same posterior, by checking the variance in the normalizing constant among runs.
tp_smc_convergence(output_list)
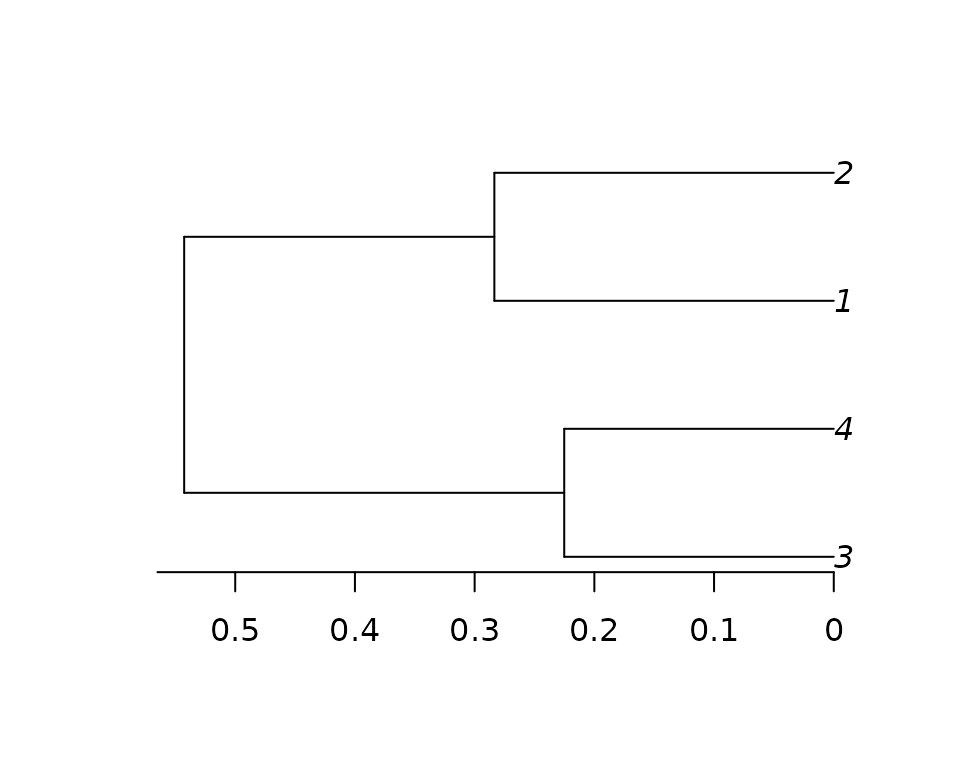
#> [1] 0.01258822There are different ways to summarize the posterior distribution of
trees. In this examples, we calculate the the MAP (Maximum A Posteriori)
tree. We do this by finding the single tree topology that has the
highest posterior probability, and then using
ape::consensus to compute average branch lengths for all
sampled trees with the MAP topology.
out_trees <- tp_json_to_phylo(output_list)
map_tree <- tp_map_tree(out_trees)
#> [1] "MAP Topology found"
#> [1] "Posterior Probability: 0.8164"
#> [1] "Based on the topology of 83 samples out of 100"
plot(map_tree)
axisPhylo()